Note
Go to the end to download the full example code.
K-Means Clustering of MNIST Dataset#
Example showing how you can perform K-Means clustering on the MNIST dataset.
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/pygfx/objects/_ruler.py:267: RuntimeWarning: divide by zero encountered in divide
screen_full = (ndc_full[:, :2] / ndc_full[:, 3:4]) * half_canvas_size
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/pygfx/objects/_ruler.py:267: RuntimeWarning: invalid value encountered in divide
screen_full = (ndc_full[:, :2] / ndc_full[:, 3:4]) * half_canvas_size
/opt/hostedtoolcache/Python/3.12.9/x64/lib/python3.12/site-packages/pygfx/objects/_ruler.py:279: RuntimeWarning: invalid value encountered in divide
screen_sel = (ndc_sel[:, :2] / ndc_sel[:, 3:4]) * half_canvas_size
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/_features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
# test_example = false
import fastplotlib as fpl
import numpy as np
from sklearn.datasets import load_digits
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
# load the data
mnist = load_digits()
# get the data and labels
data = mnist['data'] # (1797, 64)
labels = mnist['target'] # (1797,)
# visualize the first 5 digits
# NOTE: this is just to give a sense of the dataset if you are unfamiliar,
# the more interesting visualization is below :D
fig_data = fpl.Figure(shape=(1, 5), size=(900, 300))
# iterate through each subplot
for i, subplot in enumerate(fig_data):
# reshape each image to (8, 8)
subplot.add_image(data[i].reshape(8,8), cmap="gray", interpolation="linear")
# add the label as a title
subplot.set_title(f"Label: {labels[i]}")
# turn off the axes and toolbar
subplot.axes.visible = False
subplot.toolbar = False
fig_data.show()
# project the data from 64 dimensions down to the number of unique digits
n_digits = len(np.unique(labels)) # 10
reduced_data = PCA(n_components=n_digits).fit_transform(data) # (1797, 10)
# performs K-Means clustering, take the best of 4 runs
kmeans = KMeans(n_clusters=n_digits, n_init=4)
# fit the lower-dimension data
kmeans.fit(reduced_data)
# get the centroids (center of the clusters)
centroids = kmeans.cluster_centers_
# plot the kmeans result and corresponding original image
figure = fpl.Figure(
shape=(1,2),
size=(700, 400),
cameras=["3d", "2d"],
controller_types=[["fly", "panzoom"]]
)
# set the axes to False
figure[0, 0].axes.visible = False
figure[0, 1].axes.visible = False
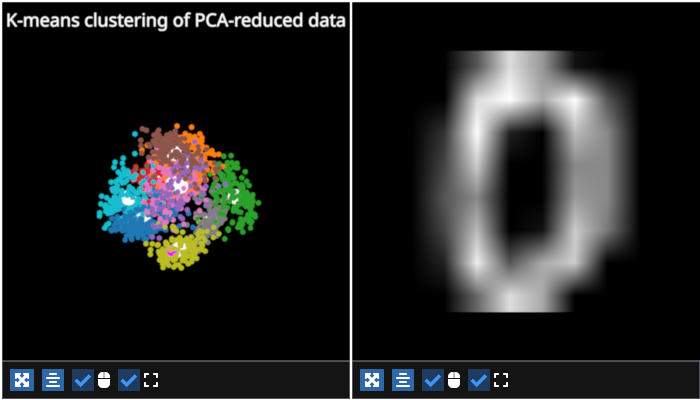
figure[0, 0].set_title(f"K-means clustering of PCA-reduced data")
# plot the centroids
figure[0, 0].add_scatter(
data=np.vstack([centroids[:, 0], centroids[:, 1], centroids[:, 2]]).T,
colors="white",
sizes=15
)
# plot the down-projected data
digit_scatter = figure[0,0].add_scatter(
data=np.vstack([reduced_data[:, 0], reduced_data[:, 1], reduced_data[:, 2]]).T,
sizes=5,
cmap="tab10", # use a qualitative cmap
cmap_transform=kmeans.labels_, # color by the predicted cluster
)
# initial index
ix = 0
# plot the initial image
digit_img = figure[0, 1].add_image(
data=data[ix].reshape(8,8),
cmap="gray",
name="digit",
interpolation="linear"
)
# change the color and size of the initial selected data point
digit_scatter.colors[ix] = "magenta"
digit_scatter.sizes[ix] = 10
# define event handler to update the selected data point
@digit_scatter.add_event_handler("pointer_enter")
def update(ev):
# reset colors and sizes
digit_scatter.cmap = "tab10"
digit_scatter.sizes = 5
# update with new seleciton
ix = ev.pick_info["vertex_index"]
digit_scatter.colors[ix] = "magenta"
digit_scatter.sizes[ix] = 10
# update digit fig
figure[0, 1]["digit"].data = data[ix].reshape(8, 8)
figure.show()
# NOTE: `if __name__ == "__main__"` is NOT how to use fastplotlib interactively
# please see our docs for using fastplotlib interactively in ipython and jupyter
if __name__ == "__main__":
print(__doc__)
fpl.loop.run()
Total running time of the script: (0 minutes 0.828 seconds)
