Note
Go to the end to download the full example code.
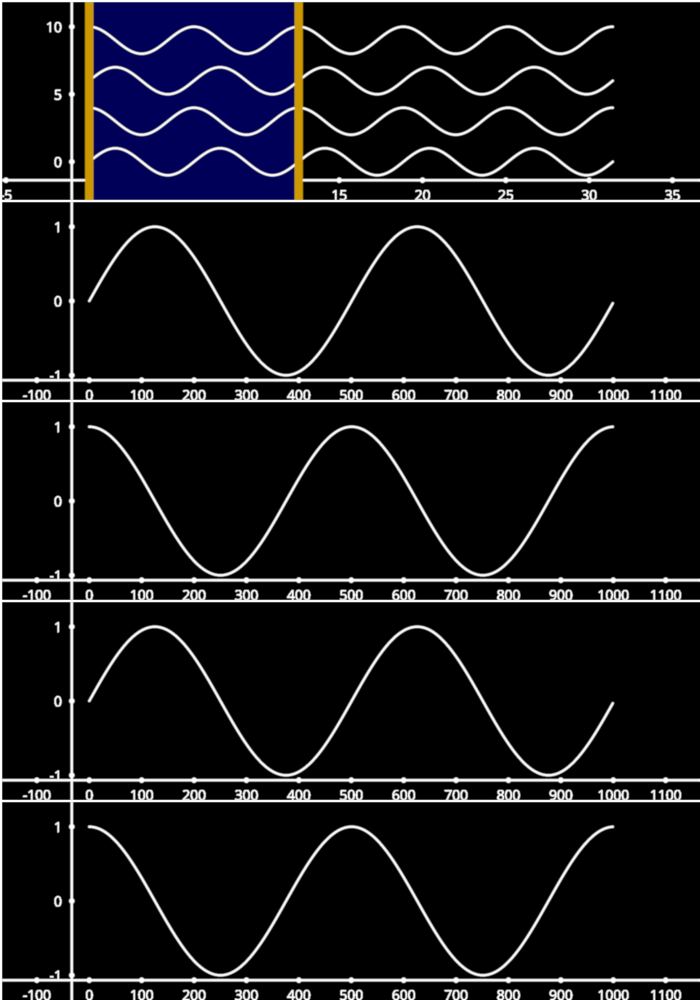
LinearRegionSelectors with LineCollection#
Example showing how to use a LinearRegionSelector with a LineCollection
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/_features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
# test_example = false
import fastplotlib as fpl
import numpy as np
# data to plot
xs = np.linspace(0, 10 * np.pi, 1_000)
sine = np.column_stack([xs, np.sin(xs)])
cosine = np.column_stack([xs, np.cos(xs)])
figure = fpl.Figure((5, 1), size=(700, 1000))
# preallocated size for zoomed data
zoomed_prealloc = 1_000
# sines and cosines
data = [sine, cosine, sine, cosine]
# make line stack
line_stack = figure[0, 0].add_line_stack(data, separation=2)
# make selector
selector = line_stack.add_linear_region_selector()
# preallocate array for storing zoomed in data
zoomed_init = np.column_stack([np.arange(zoomed_prealloc), np.zeros(zoomed_prealloc)])
# populate zoomed view subplots with graphics using preallocated buffer sizes
for i, subplot in enumerate(figure):
if i == 0:
# skip the first one
continue
# make line graphics for displaying zoomed data
subplot.add_line(zoomed_init, name="zoomed")
def interpolate(subdata: np.ndarray, axis: int):
"""1D interpolation to display within the preallocated data array"""
x = np.arange(0, zoomed_prealloc)
xp = np.linspace(0, zoomed_prealloc, subdata.shape[0])
# interpolate to preallocated size
return np.interp(x, xp, fp=subdata[:, axis]) # use the y-values
@selector.add_event_handler("selection")
def update_zoomed_subplots(ev):
"""update the zoomed subplots"""
zoomed_data = ev.get_selected_data()
for i in range(len(zoomed_data)):
# interpolate y-vals
data = interpolate(zoomed_data[i], axis=1)
figure[i + 1, 0]["zoomed"].data[:, 1] = data
figure[i + 1, 0].auto_scale()
# set initial selection so zoomed plots update
selector.selection = (0, 4 * np.pi)
# hide toolbars to reduce clutter
for subplot in figure:
subplot.toolbar = False
figure.show(maintain_aspect=False)
# NOTE: `if __name__ == "__main__"` is NOT how to use fastplotlib interactively
# please see our docs for using fastplotlib interactively in ipython and jupyter
if __name__ == "__main__":
print(__doc__)
fpl.run()
Total running time of the script: (0 minutes 2.100 seconds)
