Note
Go to the end to download the full example code.
LinearRegionSelectors#
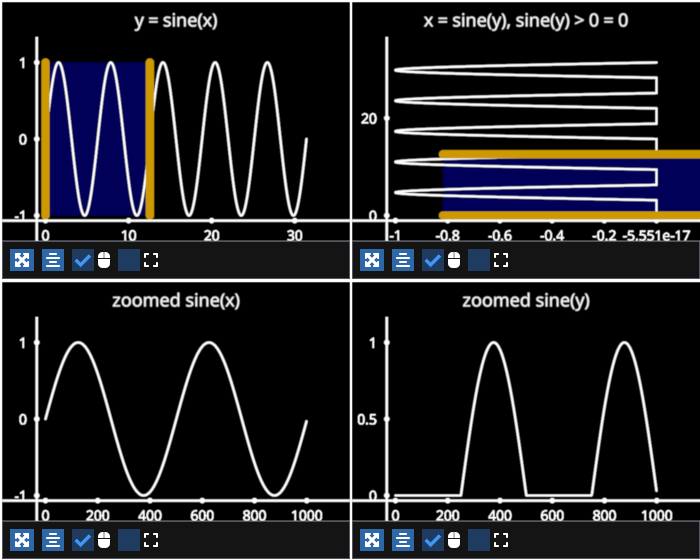
Example showing how to use a LinearRegionSelector with lines. We demonstrate two use cases, a horizontal LinearRegionSelector which selects along the x-axis and a vertical selector which moves along the y-axis.
In general, a horizontal selector on the x-axis is useful if you are displaying data where y = f(x). Conversely, a vertical selector that selectors along the y-axis is useful for displaying data where x = f(y). (ex: vertical histograms)
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/_features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
# test_example = false
import fastplotlib as fpl
import numpy as np
# names for out subplots
names = [
["y = sine(x)", "x = sine(y), sine(y) > 0 = 0"],
["zoomed sine(x)", "zoomed sine(y)"]
]
# 2 rows, 2 columns
figure = fpl.Figure(
(2, 2),
size=(700, 560),
names=names,
)
# preallocated size for zoomed data
zoomed_prealloc = 1_000
# data to plot
xs = np.linspace(0, 10 * np.pi, 1_000)
ys = np.sin(xs) # y = sine(x)
# make sine along x axis
sine_graphic_x = figure[0, 0].add_line(np.column_stack([xs, ys]))
# x = sine(y), sine(y) > 0 = 0
sine_y = ys
sine_y[sine_y > 0] = 0
# sine along y axis
sine_graphic_y = figure[0, 1].add_line(np.column_stack([ys, xs]))
# offset the position of the graphic to demonstrate `get_selected_data()` later
sine_graphic_y.position_x = 50
sine_graphic_y.position_y = 50
# add linear selectors
selector_x = sine_graphic_x.add_linear_region_selector() # default axis is "x"
selector_y = sine_graphic_y.add_linear_region_selector(axis="y")
# preallocate array for storing zoomed in data
zoomed_init = np.column_stack([np.arange(zoomed_prealloc), np.zeros(zoomed_prealloc)])
# make line graphics for displaying zoomed data
zoomed_x = figure[1, 0].add_line(zoomed_init)
zoomed_y = figure[1, 1].add_line(zoomed_init)
def interpolate(subdata: np.ndarray, axis: int):
"""1D interpolation to display within the preallocated data array"""
x = np.arange(0, zoomed_prealloc)
xp = np.linspace(0, zoomed_prealloc, subdata.shape[0])
# interpolate to preallocated size
return np.interp(x, xp, fp=subdata[:, axis]) # use the y-values
@selector_x.add_event_handler("selection")
def set_zoom_x(ev):
"""sets zoomed x selector data"""
# get the selected data
selected_data = ev.get_selected_data()
if selected_data.size == 0:
# no data selected
zoomed_x.data[:, 1] = 0
# interpolate the y-values since y = f(x)
zoomed_x.data[:, 1] = interpolate(selected_data, axis=1)
figure[1, 0].auto_scale()
def set_zoom_y(ev):
"""sets zoomed x selector data"""
# get the selected data
selected_data = ev.get_selected_data()
if selected_data.size == 0:
# no data selected
zoomed_y.data[:, 1] = 0
# interpolate the x values since this x = f(y)
zoomed_y.data[:, 1] = -interpolate(selected_data, axis=0)
figure[1, 1].auto_scale()
# you can also add event handlers without a decorator
selector_y.add_event_handler(set_zoom_y, "selection")
# set initial selection
selector_x.selection = selector_y.selection = (0, 4 * np.pi)
figure.show(maintain_aspect=False)
# NOTE: `if __name__ == "__main__"` is NOT how to use fastplotlib interactively
# please see our docs for using fastplotlib interactively in ipython and jupyter
if __name__ == "__main__":
print(__doc__)
fpl.run()
Total running time of the script: (0 minutes 3.724 seconds)
