Note
Go to the end to download the full example code.
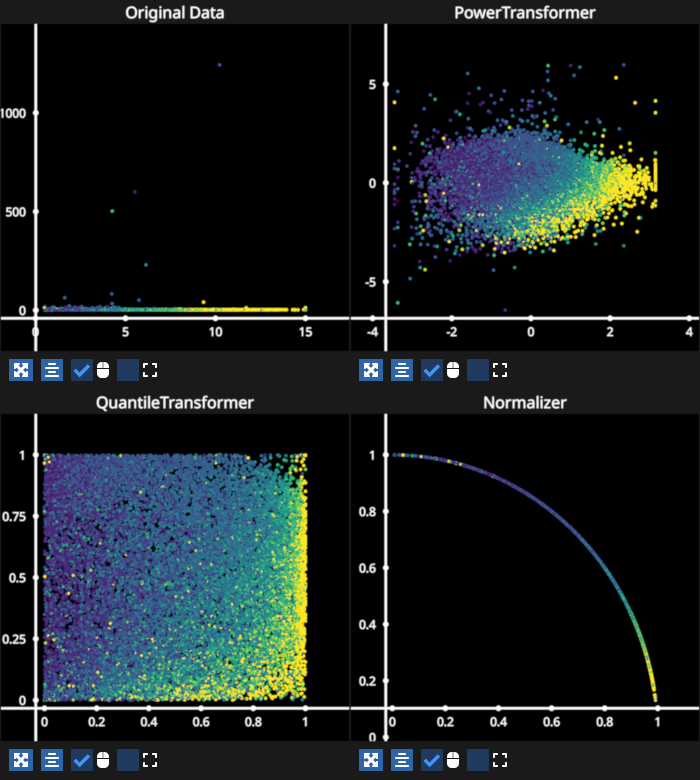
Scatter data explore scalers#
Based on the sklearn preprocessing scalers example. Hover points to highlight the corresponding point of the dataset transformed by the various scalers.
See: https://scikit-learn.org/stable/auto_examples/preprocessing/plot_all_scaling.html
This is another example that uses bi-directional events.
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/sklearn/preprocessing/_data.py:2885: UserWarning: n_quantiles (1000) is greater than the total number of samples (442). n_quantiles is set to n_samples.
warnings.warn(
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
/home/runner/work/fastplotlib/fastplotlib/fastplotlib/graphics/features/_base.py:18: UserWarning: casting float64 array to float32
warn(f"casting {array.dtype} array to float32")
# test_example = false
from sklearn.datasets import load_diabetes
from sklearn.preprocessing import (
StandardScaler,
QuantileTransformer,
PowerTransformer,
)
import fastplotlib as fpl
import pygfx
# get the dataset
dataset = load_diabetes(scaled=False)
# Take only 2 features to make visualization easier
X = dataset["data"][:, (2, 6)]
# target
y = dataset["target"]
# list of our scalers and their names as strings
scalers = [PowerTransformer, QuantileTransformer, StandardScaler]
names = ["Original Data", *[s.__name__ for s in scalers]]
# fastplotlib code starts here, make a figure
figure = fpl.Figure(
shape=(2, 2),
names=names,
size=(700, 780),
)
scatters = list() # list to store our 4 scatter graphics for convenience
# add a scatter of the original data
s = figure["Original Data"].add_scatter(
data=X,
cmap="viridis",
cmap_transform=y,
sizes=3,
)
# append to list of scatters
scatters.append(s)
# add the scaled data as scatter graphics
for scaler in scalers:
name = scaler.__name__
s = figure[name].add_scatter(scaler().fit_transform(X), cmap="viridis", cmap_transform=y, sizes=3)
scatters.append(s)
# simple dict to restore the original scatter color and size
# of the previously clicked point upon clicking a new point
old_props = {"index": None, "size": None, "color": None}
def highlight_point(ev: pygfx.PointerEvent):
# event handler to highlight the point when the mouse moves over it
global old_props
# the index of the point that was just clicked
new_index = ev.pick_info["vertex_index"]
# restore old point's properties
if old_props["index"] is not None:
old_index = old_props["index"]
if new_index == old_index:
# same point was clicked, ignore
return
for s in scatters:
s.colors[old_index] = old_props["color"]
s.sizes[old_index] = old_props["size"]
# store the current property values of this new point
old_props["index"] = new_index
# all the scatters have the same colors and size for the corresponding index
# so we can just use the first scatter's original color and size
old_props["color"] = scatters[0].colors[new_index].copy() # if you do not copy you will just get a view of the array!
old_props["size"] = scatters[0].sizes[new_index]
# highlight this new point
for s in scatters:
s.colors[new_index] = "magenta"
s.sizes[new_index] = 15
# add the event handler to all the scatter graphics
for s in scatters:
s.add_event_handler(highlight_point, "pointer_move")
figure.show(maintain_aspect=False)
# NOTE: fpl.loop.run() should not be used for interactive sessions
# See the "JupyterLab and IPython" section in the user guide
if __name__ == "__main__":
print(__doc__)
fpl.loop.run()
Total running time of the script: (0 minutes 0.728 seconds)
